ClearMap.ImageProcessing package¶
This sub-package provides routines for volumetric image processing in parallel
This part of the ClearMap toolbox is designed in a modular way to allow for fast and flexible extension and addition of specific image processing algorithms.
- The toolbox part consists of two parts:
Volumetric Image Processing¶
The image processing routines provided in the standard package are listed below
| Module | Descrition |
|---|---|
BackgroundRemoval |
Background estimation and removal via morphological opening |
IlluminationCorrection |
Correction of vignetting and other illumination errors |
GreyReconstruction |
Reconstruction of images |
Filter |
Filtering of images via a large set of filter kernels |
MaximaDetection |
Detection of maxima and h-max transforms |
SpotDetection |
Detection of local peaks / spots / nuclei |
CellDetection |
Detection of cells |
CellSizeDetection |
Detection of cell shapes and volumes via e.g. watershed |
IlastikClassification |
Classification of voxels via interface to Ilastik |
While some of these modules provide basic volumetric image processing functionality some routines combine those functions to provide predefined higher level cell detection, cell size and intensity measurements.
The higher level routines are optimized for iDISCO+ cleared mouse brain samples stained for c-Fos expression. Other data sets might require a redesign of these higher level functions.
Parallel Image Processing¶
For large volumetric image data sets from e.g. light sheet microscopy parallel processing is essential to speed up calculations.
In this toolbox the image processing is parallelized via splitting a volumetric image stack into several sub-stacks, typically in z-direction. Because most of the image processing steps are non-local sub-stacks are created with overlaps and the results rejoined accordingly to minimize boundary effects.
Parallel processing is handled via the
StackProcessing module.
External Packages¶
The ImageProcessing module makes use of external image
processing packages including:
Routines form these packages were freely chosen to optimize for speed and memory consumption
ClearMap.ImageProcessing.StackProcessing module¶
Process a image stack in parallel or sequentially
In this toolbox image processing is parallized via splitting a volumetric image stack into several sub-stacks, typically in z-direction. As most of the image processig steps are non-local sub-stacks are created with overlaps and the results rejoined accordingly to minimize boundary effects.
Parallel processing is handled via this module.
Sub-Stacks¶
The parallel processing module creates a dictionary with information on the sub-stack as follows:
| Key | Description |
|---|---|
stackId |
id of the sub-stack |
nStacks |
total number of sub-stacks |
source |
source file/folder/pattern of the stack |
x, y, z |
the range of the sub-stack with in the full image |
zCenters |
tuple of the centers of the overlaps |
zCenterIndices |
tuple of the original indices of the centers of the overlaps |
zSubStackCenterIndices |
tuple of the indices of the sub-stack that correspond to the overlap centers |
For exmaple the writeSubStack() routine makes uses of this information
to write out only the sub-parts of the image that is will contribute to the
final total image.
-
printSubStackInfo(subStack, out=<open file '<stdout>', mode 'w'>)[source]¶ Print information about the sub-stack
Parameters: - subStack (dict) – the sub-stack info
- out (object) – the object to write the information to
-
writeSubStack(filename, img, subStack=None)[source]¶ Write the non-redundant part of a sub-stack to disk
The routine is used to write out images when porcessed in parallel. It assumes that the filename is a patterned file name.
Parameters: - filename (str or None) –
file name pattern as described in
FileList, if None return as array - img (array) – image data of sub-stack
- subStack (dict or None) – sub-stack information, if None write entire image see Sub-Stacks
Returns: str or array – the file name pattern or image
- filename (str or None) –
file name pattern as described in
-
joinPoints(results, subStacks=None, shiftPoints=True, **args)[source]¶ Joins a list of points obtained from processing a stack in chunks
Parameters: - results (list) – list of point results from the individual sub-processes
- subStacks (list or None) – list of all sub-stack information, see Sub-Stacks
- shiftPoints (bool) – if True shift points to refer to origin of the image stack considered when range specification is given. If False, absolute position in entire image stack.
Returns: tuple – joined points, joined intensities
-
calculateChunkSize(size, processes=2, chunkSizeMax=100, chunkSizeMin=30, chunkOverlap=15, chunkOptimization=True, chunkOptimizationSize=<built-in function all>, verbose=True)[source]¶ Calculates the chunksize and other info for parallel processing
The sub stack information is described in Sub-Stacks
Parameters: - processes (int) – number of parallel processes
- chunkSizeMax (int) – maximal size of a sub-stack
- chunkSizeMin (int) – minial size of a sub-stack
- chunkOverlap (int) – minimal sub-stack overlap
- chunkOptimization (bool) – optimize chunck sizes to best fit number of processes
- chunkOptimizationSize (bool or all) – if True only decrease the chunk size when optimizing
- verbose (bool) – print information on sub-stack generation
Returns: tuple – number of chunks, z-ranges of each chunk, z-centers in overlap regions
-
calculateSubStacks(source, z=<built-in function all>, x=<built-in function all>, y=<built-in function all>, **args)[source]¶ Calculates the chunksize and other info for parallel processing and returns a list of sub-stack objects
The sub-stack information is described in Sub-Stacks
Parameters: - source (str) – image source
- x,y,z (tuple or all) – range specifications
- processes (int) – number of parallel processes
- chunkSizeMax (int) – maximal size of a sub-stack
- chunkSizeMin (int) – minial size of a sub-stack
- chunkOverlap (int) – minimal sub-stack overlap
- chunkOptimization (bool) – optimize chunck sizes to best fit number of processes
- chunkOptimizationSize (bool or all) – if True only decrease the chunk size when optimizing
- verbose (bool) – print information on sub-stack generation
Returns: list – list of sub-stack objects
-
noProcessing(img, **parameter)[source]¶ Perform no image processing at all and return original image
Used as the default functon in
parallelProcessStack()andsequentiallyProcessStack().Parameters: img (array) – imag Returns: (array) – the original image
-
parallelProcessStack(source, x=<built-in function all>, y=<built-in function all>, z=<built-in function all>, sink=None, processes=2, chunkSizeMax=100, chunkSizeMin=30, chunkOverlap=15, chunkOptimization=True, chunkOptimizationSize=<built-in function all>, function=<function noProcessing>, join=<function joinPoints>, verbose=False, **parameter)[source]¶ Parallel process a image stack
Main routine that distributes image processing on paralllel processes.
Parameters: - source (str) – image source
- x,y,z (tuple or all) – range specifications
- sink (str or None) – destination for the result
- processes (int) – number of parallel processes
- chunkSizeMax (int) – maximal size of a sub-stack
- chunkSizeMin (int) – minial size of a sub-stack
- chunkOverlap (int) – minimal sub-stack overlap
- chunkOptimization (bool) – optimize chunck sizes to best fit number of processes
- chunkOptimizationSize (bool or all) – if True only decrease the chunk size when optimizing
- function (function) – the main image processing script
- join (function) – the fuction to join the results from the image processing script
- verbose (bool) – print information on sub-stack generation
Returns: str or array – results of the image processing
-
sequentiallyProcessStack(source, x=<built-in function all>, y=<built-in function all>, z=<built-in function all>, sink=None, chunkSizeMax=100, chunkSizeMin=30, chunkOverlap=15, function=<function noProcessing>, join=<function joinPoints>, verbose=False, **parameter)[source]¶ Sequential image processing on a stack
Main routine that sequentially processes a large image on sub-stacks.
Parameters: - source (str) – image source
- x,y,z (tuple or all) – range specifications
- sink (str or None) – destination for the result
- processes (int) – number of parallel processes
- chunkSizeMax (int) – maximal size of a sub-stack
- chunkSizeMin (int) – minial size of a sub-stack
- chunkOverlap (int) – minimal sub-stack overlap
- chunkOptimization (bool) – optimize chunck sizes to best fit number of processes
- chunkOptimizationSize (bool or all) – if True only decrease the chunk size when optimizing
- function (function) – the main image processing script
- join (function) – the fuction to join the results from the image processing script
- verbose (bool) – print information on sub-stack generation
Returns: str or array – results of the image processing
ClearMap.ImageProcessing.CellDetection module¶
Cell Detection Module
This is the main routine to run the individual routines to detect cells om volumetric image data.
ClearMap supports two predefined image processing pipelines which will extend in the future:
| Method | Description | Reference |
|---|---|---|
| “SpotDetection” | uses predefined spot detection pipline | detectCells() |
| “Ilastik” | uses predefined pipline with cell classification via Ilastik | classifyCells() |
| function | a user defined function | NA |
Example
>>> import ClearMap.IO as io
>>> import ClearMap.Settings as settings
>>> from ClearMap.ImageProcessing.CellDetection import detectCells;
>>> fn = os.path.join(settings.ClearMapPath, 'Test/Data/Synthetic/test_iDISCO_\d{3}.tif');
>>> parameter = {"filterDoGParameter" : {"size": (5,5,5)}, "findExtendedMaximaParameter" : {"threshold" : 5}};
>>> img = io.readData(fn);
>>> img = img.astype('int16'); # converting data to smaller integer types can be more memory efficient!
>>> res = detectCells(img, parameter);
>>> print res[0].shape
See also
-
detectCells(source, sink=None, method='SpotDetection', processMethod=<built-in function all>, verbose=False, **parameter)[source]¶ Detect cells in data
This is a main script to start running the cell detection.
Parameters: source (str or array) – Image source
sink (str or None) – destination for the results
method (str or function) –
Method Description “SpotDetection” uses predefined spot detection pipline “Ilastik” uses predefined pipline with cell classification via Ilastik function a user defined function processMethod (str or all) – ‘sequential’ or ‘parallel’. if all its choosen automatically
verbose (bool) – print info
**parameter (dict) – parameter for the image procesing sub-routines
Returns:
ClearMap.ImageProcessing.CellSizeDetection module¶
Cell shape and size detection routines
The cell shape detection is based on a seeded and masked watershed.
-
detectCellShape(img, peaks, detectCellShapeParameter=None, threshold=None, save=None, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find cell shapes as labeled image
Parameters: img (array) – image data
peaks (array) – point data of cell centers / seeds
detectCellShape (dict) –
Name Type Descritption threshold (float or None) threshold to determine mask, pixel below this are background if None no mask is generated save (tuple) size of the box on which to perform the method verbose (bool or int) print / plot information about this step verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – labeled image where each label indicates a cell
-
findCellSize(imglabel, findCelSizeParameter=None, maxLabel=None, verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find cell size given cell shapes as labled image
Parameters: imglabel (array or str) – labeled image, where each cell has its own label
findCelSizeParameter (dict) –
Name Type Descritption maxLabel (int or None) maximal label to include, if None determine automatically verbose (bool or int) print / plot information about this step verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – measured intensities
-
findCellIntensity(img, imglabel, findCellIntensityParameter=None, maxLabel=None, method='Sum', verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find integrated cell intensity given cell shapes as labled image
Parameters: img (array or str) – image data
imglabel (array or str) – labeled image, where each cell has its own label
findCellIntensityParameter (dict) –
Name Type Descritption maxLabel (int or None) maximal label to include, if None determine automatically method (str) method to use for measurment: ‘Sum’, ‘Mean’, ‘Max’, ‘Min’ verbose (bool or int) print / plot information about this step verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – measured intensities
Subpackages¶
ClearMap.ImageProcessing.IlluminationCorrection module¶
Illumination correction toolbox.
The module provides a function to correct illumination/vignetting systematic variations in intensity.
The intensity image  given a flat field
given a flat field 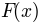 and
a background
and
a background 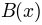 the image is corrected to
the image is corrected to 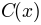 as:
as:
The module also has functionality to create flat field corections from measured
intensity changes in a single direction, useful e.g. for lightsheet images,
see e.g. flatfieldLineFromRegression().
References
Fundamentals of Light Microscopy and Electronic Imaging, p. 421
-
DefaultFlatFieldLineFile= '/home/mtllab/Programs/ClearMap/idisco/ClearMap/Data/lightsheet_flatfield_correction.csv'¶ Default file of points along the illumination changing line for the flat field correction
See also
-
correctIllumination(img, correctIlluminationParameter=None, flatfield=None, background=None, scaling=None, save=None, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Correct illumination variations
The intensity image
 given a flat field
given a flat field 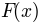 and
a background
and
a background 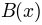 the image is corrected to
the image is corrected to 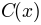 as:
as:If the background is not given
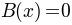 .
.The correction is done slice by slice assuming the data was collected with a light sheet microscope.
The image is finally optionally scaled.
Parameters: img (array) – image data
findCenterOfMaximaParameter (dict) –
Name Type Descritption flatfield (str, None or array) flat field intensities, if None d onot correct image for illumination, if True the background (str, None or array) background image as file name or array if None background is assumed to be zero scaling (str or None) scale the corrected result by this factor if ‘max’/’mean’ scale to keep max/mean invariant save (str or None) save the corrected image to file verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – illumination corrected image
References
Fundamentals of Light Microscopy and Electronic Imaging, p 421
See also
-
flatfieldFromLine(line, xsize)[source]¶ Creates a 2d flat field image from a 1d line of estimated intensities
Parameters: - lines (array) – array of intensities along y axis
- xsize (int) – size of image in x dimension
Returns: array – full 2d flat field
-
flatfieldLineFromRegression(data, sink=None, method='polynomial', reverse=None, verbose=False)[source]¶ Create flat field line fit from a list of positions and intensities
The fit is either to be assumed to be a Gaussian:
or follows a order 6 radial polynomial
Parameters: - data (array) – intensity data as vector of intensities or (n,2) dim array of positions d=0 and intensities measurements d=1:-1
- sink (str or None) – destination to write the result of the fit
- method (str) – method to fit intensity data, ‘Gaussian’ or ‘Polynomial’
- reverse (bool) – reverse the line fit after fitting
- verbose (bool) – print and plot information for the fit
Returns: array – fitted intensities on points
ClearMap.ImageProcessing.BackgroundRemoval module¶
Functions to remove background in images
The main routine subtracts a morphological opening from the original image for background removal.
-
removeBackground(img, removeBackgroundParameter=None, size=None, save=None, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Remove background via subtracting a morphological opening from the original image
Background removal is done z-slice by z-slice.
Parameters: img (array) – image data
removeBackGroundParameter (dict) –
Name Type Descritption size (tuple or None) size for the structure element of the morphological opening if None, do not correct for any background save (str or None) file name to save result of this operation if None dont save to file verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – background corrected image
ClearMap.ImageProcessing.GreyReconstruction module¶
Grey reconstruction module
This morphological reconstruction routine was adapted from CellProfiler.
Author
Original author: Lee Kamentsky Copyright (c) 2003-2009 Massachusetts Institute of Technology Copyright (c) 2009-2011 Broad Institute
Modified by Chirstoph Kirst to optimize integration into ClearMap, The Rockefeller University, New York City, 2015
-
reconstruct(seed, mask, method='dilation', selem=None, offset=None)[source]¶ Performs a morphological reconstruction of an image.
Reconstruction uses a seed image, which specifies the values to dilate and a mask image that gives the maximum allowed dilated value at each pixel.
The algorithm is taken from [1]. Applications for greyscale reconstruction are discussed in [2] and [3].
Parameters: - seed (array) – seed image to be dilated or eroded.
- mask (array) – maximum (dilation) / minimum (erosion) allowed
- method (str) – {‘dilation’|’erosion’}
- selem (array) – structuring element
- offset (array or None) – offset of the structuring element, None is centered
Returns: array – result of morphological reconstruction.
Note
Operates on 2d images.
Reference:
[1] Robinson, “Efficient morphological reconstruction: a downhill filter”, Pattern Recognition Letters 25 (2004) 1759-1767. [2] Vincent, L., “Morphological Grayscale Reconstruction in Image Analysis: Applications and Efficient Algorithms”, IEEE Transactions on Image Processing (1993) [3] Soille, P., “Morphological Image Analysis: Principles and Applications”, Chapter 6, 2nd edition (2003), ISBN 3540429883.
-
greyReconstruction(img, mask, greyReconstructionParameter=None, method=None, size=3, save=None, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Calculates the grey reconstruction of the image
Reconstruction is done z-slice by z-slice.
Parameters: img (array) – image data
removeBackGroundParameter (dict) –
Name Type Descritption method (tuple or None) ‘dilation’ or ‘erosion’, if None return original image size (int or tuple) size of structuring element save (str or None) file name to save result of this operation if None dont save to file verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – grey reconstructed image
ClearMap.ImageProcessing.SpotDetection module¶
Functions to detect spots in images
The main routine detectCells() uses a difference of gaussian filter (see
Filter) followed by a peak detection step.
Example
>>> import os
>>> import ClearMap.IO as io
>>> import ClearMap.Settings as settings
>>> import ClearMap.ImageProcessing.SpotDetection as sd
>>> fn = os.path.join(settings.ClearMapPath, 'Test/Data/Synthetic/test_iDISCO_\d{3}.tif');
>>> img = io.readData(fn);
>>> img = img.astype('int16'); # converting data to smaller integer types can be more memory efficient!
>>> res = sd.detectSpots(img, dogSize = (5,5,5), flatfield = None, threshold = 5, cellShapeThreshold = 1);
>>> print 'Found %d cells !' % res[0].shape[0]
Illumination: flatfield : None
Illumination: illuminationScaling: True
Illumination: background : None
Background: backgroundSize: (15, 15)
Background: elapsed time: 0:00:00
DoG: dogSize: (5, 5, 5)
DoG: elapsed time: 0:00:00
Extended Max: threshold : 5
Extended Max: localMaxSize: 5
Extended Max: hMax : None
Extended Max: elapsed time: 0:00:00
Cell Centers: elapsed time: 0:00:00
Cell Shape: cellShapeThreshold: 1
Cell Shape:: elapsed time: 0:00:00
Cell Size:: elapsed time: 0:00:00
Cell Intensity: cellIntensityMethod: Max
Cell Intensity:: elapsed time: 0:00:00
Cell Intensity: cellIntensityMethod: Max
Cell Intensity:: elapsed time: 0:00:00
Cell Intensity: cellIntensityMethod: Max
Cell Intensity:: elapsed time: 0:00:00
Found 38 cells !
After execution this example inspect the result of the cell detection in the folder ‘Test/Data/CellShape/cellshape_d{3}.tif’.
-
detectSpots(img, detectSpotsParameter=None, correctIlluminationParameter=None, removeBackgroundParameter=None, filterDoGParameter=None, findExtendedMaximaParameter=None, detectCellShapeParameter=None, verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Detect Cells in 3d grayscale image using DoG filtering and maxima detection
- Effectively this function performs the following steps:
- illumination correction via
correctIllumination() - background removal via
removeBackground() - difference of Gaussians (DoG) filter via
filterDoG() - maxima detection via
findExtendedMaxima() - cell shape detection via
detectCellShape() - cell intensity and size measurements via:
findCellIntensity(),findCellSize().
- illumination correction via
detectCells .. note:: Processing steps are done in place to save memory.
Parameters: - img (array) – image data
- detectSpotParameter – image processing parameter as described in the individual sub-routines
- verbose (bool) – print progress information
- out (object) – object to print progress information to
Returns: tuple – tuple of arrays (cell coordinates, raw intensity, fully filtered intensty, illumination and background corrected intensity [, cell size])
ClearMap.ImageProcessing.MaximaDetection module¶
Collection of routines to detect maxima
Used for finding cells or intensity peaks.
-
hMaxTransform(img, hMax)[source]¶ Calculates h-maximum transform of an image
Parameters: - img (array) – image
- hMax (float or None) – h parameter of h-max transform
Returns: array – h-max transformed image if h is not None
-
localMax(img, size=5)[source]¶ Calculates local maxima of an image
Parameters: - img (array) – image
- size (float or None) – size of volume to search for maxima
Returns: array – mask that is True at local maxima
-
extendedMax(img, hMax=0)[source]¶ Calculates extened h maxima of an image
Extended maxima are the local maxima of the h-max transform
Parameters: - img (array) – image
- hMax (float or None) – h parameter of h-max transform
Returns: array – extended maxima of the image
-
findExtendedMaxima(img, findExtendedMaximaParameter=None, hMax=None, size=5, threshold=None, save=None, verbose=None, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find extended maxima in an image
Effectively this routine performs a h-max transfrom, followed by a local maxima search and thresholding of the maxima.
Parameters: img (array) – image data
findExtendedMaximaParameter (dict) –
Name Type Descritption hMax (float or None) h parameter for the initial h-Max transform if None, do not perform a h-max transform size (tuple) size for the structure element for the local maxima filter threshold (float or None) include only maxima larger than a threshold if None keep all localmaxima save (str or None) file name to save result of this operation if None do not save result to file verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – binary image with True pixel at extended maxima
See also
-
findCenterOfMaxima(img, imgmax=None, label=None, findCenterOfMaximaParameter=None, save=None, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find center of detected maxima weighted by intensity
Parameters: img (array) – image data
findCenterOfMaximaParameter (dict) –
Name Type Descritption save (str or None) saves result of labeling the differnet maxima if None, do the lableling is not saved verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – coordinates of centers of maxima, shape is (n,d) where n is number of maxima and d the dimension of the image
-
findPixelCoordinates(imgmax, subStack=None, verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find coordinates of all pixel in an image with positive or True value
Parameters: - img (array) – image data
- verbose (bool) – print progress info
- out (object) – object to write progress info to
Returns: array – coordinates of centers of True pixels, shape is (n,d) where n is number of maxima and d the dimension of the image
-
findIntensity(img, centers, findIntensityParameter=None, method=None, size=(3, 3, 3), verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Find instensity value around centers in the image
Parameters: img (array) – image data
findIntensityParameter (dict) –
Name Type Descritption method (str, func, None) method to use to determine intensity (e.g. “Max” or “Mean”) if None take intensities at the given pixels size (tuple) size of the box on which to perform the method verbose (bool or int) print / plot information about this step verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array – measured intensities
ClearMap.ImageProcessing.IlastikClassification module¶
Inteface to Illastik pixel classification
This module allows to integrate ilastik pixel classification into the ClearMap pipeline.
To train a classifier ilastik should be used:
Note
Note that ilastik classification works in parallel, thus it is advised to
process the data sequentially, see
sequentiallyProcessStack()
Note
Ilastik 0.5 works for images in uint8 format !
References
- Ilastik
- Based on the ilastik interface from cell profiler
-
isInitialized()[source]¶ Check if Ilastik is useable
Returns: bool – True if Ilastik is installed and useable by ClearMap
-
checkInitialized()[source]¶ Checks if ilastik is initialized
Returns: bool – True if ilastik paths are set.
-
classifyPixel(img, classifyPixelParameter=None, subStack=None, verbose=False, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Detect Cells Using a trained classifier in Ilastik
- Arguments:from ClearMap.ImageProcessing.CellSizeDetection import detectCellShape, findCellSize, findCellIntensity
img (array): image data classifyPixelParameter (dict):
Name Type Descritption classifier (str or None) Ilastik project file with trained pixel classifier save (str or None) save the classification propabilities to a file verbose (bool or int) print / plot information about this step subStack (dict or None): sub-stack information verbose (bool): print progress info out (object): object to write progress info to
Returns: array – probabilities for each pixel to belong to a class in the classifier, shape is (img.shape, number of classes)
-
classifyCells(img, classifyCellsParameter=None, classifier=None, classindex=0, save=None, verbose=False, detectCellShapeParameter=None, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Detect Cells Using a trained classifier in Ilastik
The routine assumes that the first class is identifying the cells.
Parameters: img (array) – image data
classifyPixelParameter (dict) –
Name Type Descritption classifier (str or None) Ilastik project file with trained pixel classifier classindex (int) class index considered to be cells save (str or None) save the detected cell pixel to a file verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: tuple – centers of the cells, intensity measurments
Note
The routine could be potentially refined to make use of background detection in ilastik
ClearMap.ImageProcessing.ImageStatistics module¶
Functions to gather iamge statistics in large volumetric images
The main routines extract information from a large volumetric image, such as the maximum or mean.
-
calculateStatistics(source, sink=None, calculateStatisticsParameter=None, method='Max', remove=True, processMethod=<built-in function all>, verbose=False, **parameter)[source]¶ Calculate statisticsfrom image data
This is a main script to start extracting statistics of volumetric image data.
Parameters: source (str or array) – Image source
sink (str or None) – destination for the results
calculateStatisticsParameter (dict) –
Name Type Descritption method (str or function) function to extract statistic, must be trivially distributable if None, do not extract information remove (bool) remove redundant overlap verbose (bool or int) print / plot information about this step method (str or function)
processMethod (str or all) – ‘sequential’ or ‘parallel’. if all its choosen automatically
verbose (bool) – print info
**parameter (dict) – parameter for the image processing sub-routines
Returns: list of statistics
-
calculateStatisticsOnStack(img, calculateStatisticsParameter=None, method='Max', remove=True, verbose=False, subStack=None, out=<open file '<stdout>', mode 'w'>, **parameter)[source]¶ Calculate a statistics from a large volumetric image
The statistics is assumed to be trivially distributable, i.e. max or mean.
Parameters: img (array) – image data
calculateStatisticsParameter (dict) –
Name Type Descritption method (str or function) function to extract statistic, must be trivially distributable if None, do not extract information remove (bool) remove redundant overlap verbose (bool or int) print / plot information about this step subStack (dict or None) – sub-stack information
verbose (bool) – print progress info
out (object) – object to write progress info to
Returns: array or number – extracted statistics
Note
One might need to choose zero overlap in the stacks to function properly!
-
joinStatistics(results, calculateStatisticsParameter=None, method='Max', subStacks=None, **parameter)[source]¶ Joins a list of calculated statistics
Parameters: results (list) – list of statics results from the individual sub-processes
calculateStatisticsParameter (dict) –
Name Type Descritption method (str or function) function to extract statistic, must be trivially distributable if None, do not extract information subStacks (list or None) – list of all sub-stack information, see Sub-Stacks
Returns: list or object – joined statistics