ClearMap.Alignment package¶
This sub-package provides an interface to alignment tools in order to register cleared samples to atlases or reference samples.
Supported functionality:
- resampling and reorientation of large volumetric images in the
Resamplingmodule.- registering volumetric data onto references via Elastix in the
Elastixmodule.
Main routines for resampling are:
resampleData()
and resamplePoints().
Main routines for elastix registration are:
alignData(),
transformData() and
transformPoints().
ClearMap.Alignment.Elastix module¶
Interface to Elastix for alignment of volumetric data
The elastix documentation can be found here.
In essence, a transformation  is sought so that for a fixed image
is sought so that for a fixed image
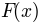 and a moving image
and a moving image  :
:

Once the map  is estimated via elastix, transformix maps an image
is estimated via elastix, transformix maps an image
 from the moving image frame to the fixed image frame, i.e.:
from the moving image frame to the fixed image frame, i.e.:

To register an image onto a reference image, the fixed image is typically choosed to be the image to be registered, while the moving image is the reference image. In this way an object identified in the data at position x is mapped via transformix as:

Summary¶
- elastix finds a transformation
- the fixed image is image to be registered
- the moving image is typically the reference image
- the result folder may contain an image (mhd file) that is
, i.e. has the size of the fixed image
- transformix applied to data gives
!
- transformix applied to points gives
!
- point arrays are assumed to be in (x,y,z) coordinates consistent with (x,y,z) array represenation of images in ClearMap
Main routines are: alignData(), transformData() and transformPoints().
See also
-
ElastixBinary= '/usr/local/elastix/bin/elastix'¶ str: the elastix executable
Notes
- setup in
initializeElastix()
- setup in
-
ElastixLib= '/usr/local/elastix/lib'¶ str: path to the elastix library
Notes
- setup in
initializeElastix()
- setup in
-
TransformixBinary= '/usr/local/elastix/bin/transformix'¶ str: the transformix executable
Notes
- setup in
initializeElastix()
- setup in
-
Initialized= True¶ bool: True if the elastixs binarys and paths are setup
Notes
- setup in
initializeElastix()
- setup in
-
setElastixLibraryPath(path=None)[source]¶ Add elastix library path to the LD_LIBRARY_PATH variable in linux
Parameters: path (str or None) – path to elastix root directory if None ClearMap.Settings.ElastixPathis used.
-
initializeElastix(path=None)[source]¶ Initialize all paths and binaries of elastix
Parameters: - path (str or None) – path to elastix root directory, if None
- :const:`ClearMap.Settings.ElastixPath` is used.
-
checkElastixInitialized()[source]¶ Checks if elastix is initialized
Returns: bool – True if elastix paths are set.
-
getTransformParameterFile(resultdir)[source]¶ Finds and returns the transformation parameter file generated by elastix
Notes
In case of multiple transformation parameter files the top level file is returned
Parameters: resultdir (str) – path to directory of elastix results Returns: str – file name of the first transformation parameter file
-
setPathTransformParameterFiles(resultdir)[source]¶ Replaces relative with abolsute path in the parameter files in the result directory
Notes
When elastix is not run in the directory of the transformation files the aboslute path needs to be given in each transformation file to point to the subsequent transformation files. This is done via this routine
Parameters: resultdir (str) – path to directory of elastix results
-
parseElastixOutputPoints(filename, indices=True)[source]¶ Parses the output points from the output file of transformix
Parameters: - filename (str) – file name of the transformix output file
- indices (bool) – if True return pixel indices otherwise float coordinates
Returns: points (array) – the transformed coordinates
-
getTransformFileSizeAndSpacing(transformfile)[source]¶ Parse the image size and spacing from a transformation parameter file
Parameters: transformfile (str) – File name of the transformix parameter file. Returns: (float, float) – the image size and spacing
-
getResultDataFile(resultdir)[source]¶ Returns the mhd result file in a result directory
Parameters: resultdir (str) – Path to elastix result directory. Returns: str – The mhd file in the result directory.
-
setTransformFileSizeAndSpacing(transformfile, size, spacing)[source]¶ Replaces size and scale in the transformation parameter file
Parameters: - transformfile (str) – transformation parameter file
- size (tuple) – the new image size
- spacing (tuplr) – the new image spacing
-
rescaleSizeAndSpacing(size, spacing, scale)[source]¶ Rescales the size and spacing
Parameters: - size (tuple) – image size
- spacing (tuple) – image spacing
- scale (tuple) – the scale factor
Returns: (tuple, tuple) – new size and spacing
-
alignData(fixedImage, movingImage, affineParameterFile, bSplineParameterFile=None, resultDirectory=None)[source]¶ Align images using elastix, estimates a transformation
 fixed image
fixed image  moving image.
moving image.Parameters: - fixedImage (str) – image source of the fixed image (typically the reference image)
- movingImage (str) – image source of the moving image (typically the image to be registered)
- affineParameterFile (str or None) – elastix parameter file for the primary affine transformation
- bSplineParameterFile (str or None) – elastix parameter file for the secondary non-linear transformation
- resultDirectory (str or None) – elastic result directory
Returns: str – path to elastix result directory
-
transformData(source, sink=[], transformParameterFile=None, transformDirectory=None, resultDirectory=None)[source]¶ Transform a raw data set to reference using the elastix alignment results
If the map determined by elastix is
 ,
transformix on data works as
,
transformix on data works as  .
.Parameters: - source (str or array) – image source to be transformed
- sink (str, [] or None) – image sink to save transformed image to. if [] return the default name of the data file generated by transformix.
- transformParameterFile (str or None) – parameter file for the primary transformation, if None, the file is determined from the transformDirectory.
- transformDirectory (str or None) – result directory of elastix alignment, if None the transformParameterFile has to be given.
- resultDirectory (str or None) – the directorty for the transformix results
Returns: array or str – array or file name of the transformed data
-
deformationField(sink=[], transformParameterFile=None, transformDirectory=None, resultDirectory=None)[source]¶ Create the deformation field T(x) - x
The map determined by elastix is

Parameters: - sink (str, [] or None) – image sink to save the transformation field; if [] return the default name of the data file generated by transformix.
- transformParameterFile (str or None) – parameter file for the primary transformation, if None, the file is determined from the transformDirectory.
- transformDirectory (str or None) – result directory of elastix alignment, if None the transformParameterFile has to be given.
- resultDirectory (str or None) – the directorty for the transformix results
Returns: array or str – array or file name of the transformed data
-
deformationDistance(deformationField, sink=None, scale=None)[source]¶ Compute the distance field from a deformation vector field
Parameters: - deformationField (str or array) –
source of the deformation field determined by
deformationField() - sink (str or None) – image sink to save the deformation field to
- scale (tuple or None) – scale factor for each dimension, if None = (1,1,1)
Returns: array or str – array or file name of the transformed data
- deformationField (str or array) –
source of the deformation field determined by
-
writePoints(filename, points, indices=True)[source]¶ Write points as elastix/transformix point file
Parameters: - filename (str) – file name of the elastix point file.
- points (array or str) – source of the points.
- indices (bool) – write as pixel indices or physical coordiantes
Returns: str – file name of the elastix point file
-
transformPoints(source, sink=None, transformParameterFile=None, transformDirectory=None, indices=True, resultDirectory=None, tmpFile=None)[source]¶ Transform coordinates math:x via elastix estimated transformation to

Note the transformation is from the fixed image coorindates to the moving image coordiantes.
Parameters: - source (str) – source of the points
- sink (str or None) – sink for transformed points
- transformParameterFile (str or None) – parameter file for the primary transformation, if None, the file is determined from the transformDirectory.
- transformDirectory (str or None) – result directory of elastix alignment, if None the transformParameterFile has to be given.
- indices (bool) – if True use points as pixel coordinates otherwise spatial coordinates.
- resultDirectory (str or None) – elastic result directory
- tmpFile (str or None) – file name for the elastix point file.
Returns: array or str – array or file name of transformed points
ClearMap.Alignment.Resampling module¶
The Resampling module provides methods to resample and reorient volumetric and point data.
Resampling the data is usually necessary as the first step to match the resolution and orientation of the reference object.
Main routines for resampling are: resampleData()
and resamplePoints().
Image Representation and Size¶
The module assumes that images in arrays are arranged as
- [x,y] or
- [x,y,z]
where x,y,z correspond to the x,y,z coordinates as displayed in e.g. ImageJ.
For example an image of size (512,512) stored in an array img will have:
>>> img.shape
(512,512)
Points are assumed to be given as x,y,z coordinates
Parameters such as resolution or dataSize are assumed to be given in (x,y) or (x,y,z) format, e.g.
>>> dataSize = (512,512)
Orientation¶
The orientation parameter is a tuple of d numbers from 1 to d that specifies the permutation of the axes, a minus sign infront of a numbeer indicates inversion of that axes. For exmaple
>>> orientation=(2,-1)
indicates that x and y should be exchanged and the new y axes should be reversed.
Generally a re-orientation is composed of first a permutation of the axes and then inverting the indicated axes.
A permutation is an orientation without signs and with numbers from 0 to d-1.
Examples
>>> import os
>>> import ClearMap.IO as io
>>> from ClearMap.Settings import ClearMapPath
>>> from ClearMap.Alignment.Resampling import resampleData
>>> filename = os.path.join(ClearMapPath,'Test/Data/OME/16-17-27_0_8X-s3-20HF_UltraII_C00_xyz-Table Z\d{4}.ome.tif');
>>> print io.dataSize(filename)
(2160, 2560, 21)
>>> data = resampleData(filename, sink = None, resolutionSource = (1,1,1), orientation = (1,2,3), resolutionSink = (10,10,2));
>>> print data.shape
(216, 256, 10)
-
fixOrientation(orientation)[source]¶ Convert orientation to standard format number sequence
Parameters: orientation (tuple or str) – orientation specification Returns: tuple – orientation sequence See also
-
inverseOrientation(orientation)[source]¶ Returns the inverse permuation of the permutation orientation taking axis inversions into account.
Parameters: orientation (tuple or str) – orientation specification Returns: tuple – orientation sequence See also
-
orientationToPermuation(orientation)[source]¶ Extracts the permuation from an orientation.
Parameters: orientation (tuple or str) – orientation specification Returns: tuple – premutation sequence See also
-
orientResolution(resolution, orientation)[source]¶ Permutes a resolution tuple according to the given orientation.
Parameters: - resolution (tuple) – resolution specification
- orientation (tuple or str) – orientation specification
Returns: tuple – oriented resolution sequence
See also
-
orientResolutionInverse(resolution, orientation)[source]¶ Permutes a resolution tuple according to the inverse of a given orientation.
Parameters: - resolution (tuple) – resolution specification
- orientation (tuple or str) – orientation specification
Returns: tuple – oriented resolution sequence
See also
-
orientDataSize(dataSize, orientation)[source]¶ Permutes a data size tuple according to the given orientation.
Parameters: - dataSize (tuple) – resolution specification
- orientation (tuple or str) – orientation specification
Returns: tuple – oriented dataSize sequence
See also
-
orientDataSizeInverse(dataSize, orientation)[source]¶ Permutes a dataSize tuple according to the inverse of a given orientation.
Parameters: - dataSize (tuple) – dataSize specification
- orientation (tuple or str) – orientation specification
Returns: tuple – oriented dataSize sequence
See also
-
resampleDataSize(dataSizeSource, dataSizeSink=None, resolutionSource=None, resolutionSink=None, orientation=None)[source]¶ Calculate scaling factors and data sizes for resampling.
Parameters: - dataSizeSource (tuple) – data size of the original image
- dataSizeSink (tuple or None) – data size of the resmapled image
- resolutionSource (tuple or None) – resolution of the source image
- resolutionSink (tuple or None) – resolution of the sink image
- orientation (tuple or str) – re-orientation specification
Returns: tuple – data size of the source tuple: data size of the sink tuple: resolution of source tuple: resolution of sink
See also
-
fixInterpolation(interpolation)[source]¶ Converts interpolation given as string to cv2 interpolation object
Parameters: interpolation (str or object) – interpolation string or cv2 object Returns: object – cv2 interpolation type
-
resampleXY(source, dataSizeSink, sink=None, interpolation='linear', out=<open file '<stdout>', mode 'w'>, verbose=True)[source]¶ Resample a 2d image slice
This routine is used for resampling a large stack in parallel in xy or xz direction.
Parameters: - source (str or array) – 2d image source
- dataSizeSink (tuple) – size of the resmapled image
- sink (str or None) – location for the resmapled image
- interpolation (str) – interpolation method to use: ‘linear’ or None (nearest pixel)
- out (stdout) – where to write progress information
- vebose (bool) – write progress info if true
Returns: array or str – resampled data or file name
-
resampleData(source, sink=None, orientation=None, dataSizeSink=None, resolutionSource=(4.0625, 4.0625, 3), resolutionSink=(25, 25, 25), processingDirectory=None, processes=1, cleanup=True, verbose=True, interpolation='linear', **args)[source]¶ Resample data of source in resolution and orientation
Parameters: - source (str or array) – image to be resampled
- sink (str or None) – destination of resampled image
- orientation (tuple) – orientation specified by permuation and change in sign of (1,2,3)
- dataSizeSink (tuple or None) – target size of the resampled image
- resolutionSource (tuple) – resolution of the source image (in length per pixel)
- resolutionSink (tuple) – resolution of the resampled image (in length per pixel)
- processingDirectory (str or None) – directory in which to perform resmapling in parallel, None a temporary directry will be created
- processes (int) – number of processes to use for parallel resampling
- cleanup (bool) – remove temporary files
- verbose (bool) – display progress information
- interpolation (str) – method to use for interpolating to the resmapled image
Returns: (array or str) – data or file name of resampled image
Notes
- resolutions are assumed to be given for the axes of the intrinsic orientation of the data and reference as when viewed by matplotlib or ImageJ
- orientation: permuation of 1,2,3 with potential sign, indicating which axes map onto the reference axes, a negative sign indicates reversal of that particular axes
- only a minimal set of information to detremine the resampling parameter has to be given, e.g. dataSizeSource and dataSizeSink
-
resampleDataInverse(sink, source=None, dataSizeSource=None, orientation=None, resolutionSource=(4.0625, 4.0625, 3), resolutionSink=(25, 25, 25), processingDirectory=None, processes=1, cleanup=True, verbose=True, interpolation='linear', **args)[source]¶ Resample data inversely to
resampleData()routineParameters: - sink (str or None) –
image to be inversly resampled (=sink in
resampleData()) - source (str or array) –
destination for inversly resmapled image (=source in
resampleData()) - dataSizeSource (tuple or None) – target size of the resampled image
- orientation (tuple) – orientation specified by permuation and change in sign of (1,2,3)
- resolutionSource (tuple) – resolution of the source image (in length per pixel)
- resolutionSink (tuple) – resolution of the resampled image (in length per pixel)
- processingDirectory (str or None) – directory in which to perform resmapling in parallel, None a temporary directry will be created
- processes (int) – number of processes to use for parallel resampling
- cleanup (bool) – remove temporary files
- verbose (bool) – display progress information
- interpolation (str) – method to use for interpolating to the resmapled image
Returns: (array or str) – data or file name of resampled image
Notes
- resolutions are assumed to be given for the axes of the intrinsic orientation of the data and reference as when viewed by matplotlib or ImageJ
- orientation: permuation of 1,2,3 with potential sign, indicating which axes map onto the reference axes, a negative sign indicates reversal of that particular axes
- only a minimal set of information to detremine the resampling parameter has to be given, e.g. dataSizeSource and dataSizeSink
- sink (str or None) –
image to be inversly resampled (=sink in
-
resamplePoints(pointSource, pointSink=None, dataSizeSource=None, dataSizeSink=None, orientation=None, resolutionSource=(4.0625, 4.0625, 3), resolutionSink=(25, 25, 25), **args)[source]¶ Resample Points to map from original data to the coordinates of the resampled image
The resampling of points here corresponds to he resampling of an image in
resampleData()Parameters: - pointSource (str or array) – image to be resampled
- pointSink (str or None) – destination of resampled image
- orientation (tuple) – orientation specified by permuation and change in sign of (1,2,3)
- dataSizeSource (str, tuple or None) – size of the data source
- dataSizeSink (str, tuple or None) – target size of the resampled image
- resolutionSource (tuple) – resolution of the source image (in length per pixel)
- resolutionSink (tuple) – resolution of the resampled image (in length per pixel)
Returns: (array or str) – data or file name of resampled points
Notes
- resolutions are assumed to be given for the axes of the intrinsic orientation of the data and reference as when viewed by matplotlib or ImageJ
- orientation: permuation of 1,2,3 with potential sign, indicating which axes map onto the reference axes, a negative sign indicates reversal of that particular axes
- only a minimal set of information to detremine the resampling parameter has to be given, e.g. dataSizeSource and dataSizeSink
-
resamplePointsInverse(pointSource, pointSink=None, dataSizeSource=None, dataSizeSink=None, orientation=None, resolutionSource=(4.0625, 4.0625, 3), resolutionSink=(25, 25, 25), **args)[source]¶ Resample points from the coordinates of the resampled image to the original data
The resampling of points here corresponds to he resampling of an image in
resampleDataInverse()Parameters: - pointSource (str or array) – image to be resampled
- pointSink (str or None) – destination of resampled image
- orientation (tuple) – orientation specified by permuation and change in sign of (1,2,3)
- dataSizeSource (str, tuple or None) – size of the data source
- dataSizeSink (str, tuple or None) – target size of the resampled image
- resolutionSource (tuple) – resolution of the source image (in length per pixel)
- resolutionSink (tuple) – resolution of the resampled image (in length per pixel)
Returns: (array or str) – data or file name of inversely resampled points
Notes
- resolutions are assumed to be given for the axes of the intrinsic orientation of the data and reference as when viewed by matplotlib or ImageJ
- orientation: permuation of 1,2,3 with potential sign, indicating which axes map onto the reference axes, a negative sign indicates reversal of that particular axes
- only a minimal set of information to detremine the resampling parameter has to be given, e.g. dataSizeSource and dataSizeSink